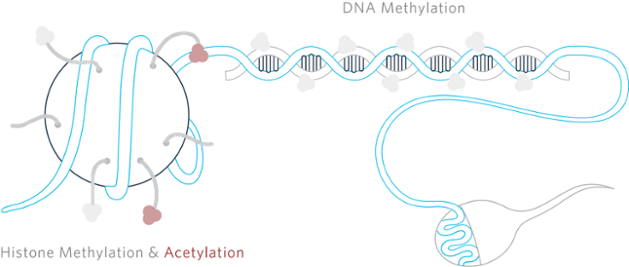
When Conrad Waddington coined the term “epigenetics” in the early 1940s, the name was used to explain why genetic variations often did not lead to phenotypic variations and how genes might actually also interact with their environment to yield a phenotype.[1] Since then there has been increasing debate and skepticism leading to polarization within the scientific research community regarding how these epigenetic changes could be passed through multiple generations without any changes occurring in the DNA.
That there are questions and challenges to epigenetics is nothing new since this is the way that new ideas usually are vetted before finally being either partially or fully embraced. However, despite this, there are those who see what can be done with the knowledge we do have as it emerges from the research.
This blog post intends to discuss both sides of the debate over the utility of epigenetics, as well as what has become of currently available applied research regarding diagnostics and the availability of new treatments emerging from epigenetics.
What is Causing the Debate?
Acceptance of Epigenetics
One of the criticisms of epigenetics is that the definition is vague and has become an all-encompassing collection of concepts, including anything that is not able to be otherwise explained: “gene regulation, physiological adaptation, disease responses…they all fall into the catch-all of epigenetics.”[2] Detractors make note of the fact that those who study epigenetics can’t even agree on common vocabulary and definitions. They also add that the epigenetic experiments have been weakly designed and there are many alternative explanations for the results, other than epigenetics[3].
However, all of this criticism obscures the fact that there are many examples of well-designed experiments that have resulted in a wealth of information. For now, the field of epigenetics “is like where genetics was 20 years ago,” says Jones, a pre-eminent leader in epigenetics. “Now it’s time to ramp it up, to move from one gene at a time to all of the genome at one time, studying all the cells to figure out how the whole thing fits together and works.”[4]
The Mechanism of Epigenetic Change
How do you explain how one cell develops into a complete human being with all the expected milestones that occur during development? Going deeper, how can virtually all epigenetic information be removed from the head of a sperm except for 10% of the original histones[5], yet what appears to be epigenetic inheritance occurs? What is the mechanism of transgenerational inheritance? How is it that most cells can revert back to the stem cell state, i.e. blank slate and develop into other tissues? The exciting part about epigenetics is that many diseases are known to have a genetic component, but the epigenetic mechanisms underlying many conditions are still being discovered with the expectation that this will lead to targeted treatments.
How are Epigenetic Changes Made Heritable?
A growing body of evidence is finding that DNA mutations, can only explain a minority of observed transmitted traits; the rest appears to be related to epigenetics. An example of epigenetic effects is imprinting.[6] Mammals inherit a set of chromosomes from each parent. Most autosomal genes will be expressed from both the maternal and the paternal alleles. Imprinted genes, however, are expressed from only one chromosome, in a parent-of-origin–dependent manner supporting the contention that epigenetics must play some active role that we have yet to understand.
The actual process of transmitting these traits along with their respective markers is not as easy as it sounds and raises more questions than answers. In order for imprinted genes to be inherited, there are a number of hurdles: 1) the chromosome must be marked as to its parental origin prior to fusion of the egg and sperm, while the maternal and paternal chromosomes are still physically separate; 2) the mark must be able to be erased and reset as development begins (most methylation is stripped from the sperm DNA at this point, although methylation remains detectable at several notable features)[7]; 3) as the cells divide and differentiate, this mark must persist; 4) a parent-of-origin mark must be recognized so that the appropriate gene is expressed In order for this information to survive through multiple generations, the signal needs to survive multiple rounds of stripping of methylation, histone, and RNA associations with DNA. As differentiation proceeds, methylation and other regulators are gradually reestablished by an as yet unknown mechanism.
What Epigenetics Can Tell Us
Central Dogma
In 1957, Francis Crick[8] outlined a startling vision of life. The ‘central dogma’ of molecular biology holds that the DNA message is transcribed into RNA which serves as a template for protein production. One gene, one protein, one tidy organization. Except that his contention doesn’t account for certain repeated observations from a growing body of scientific and medical disciplines that is not consistent with the “central dogma”. For example, it is widely recognized that identical twins with identical DNA can have dramatically different phenotypes. Another example is the study that showed that pregnant women exposed to famine in late pregnancy gave birth to smaller babies who had an increased risk of insulin resistance, obesity, dyslipidemia, and coronary heart disease.[9]
Slowly, the concept of the epigenome is being embraced. This brings up several dichotomies: Nature (genes) versus Nurture (experience with the environment), genome and epigenome as separate entities, evolution versus programming. While small changes in DNA sequence over long periods of time enhance some adaptations, this behavior does not cover the swift changes in the expression DNA that are required with environmental changes. Epigenetic factors appear to be more important than the influences of genomic information, Genetic factors account for only 20% of the heritability of diseases such as Crohn’s[10] implying that epigenetic factors are far more important than what the genetic code is. Genes appear to link evolution and development in a number of ways.
As contentious as it sounds, both the central dogma and epigenetics can not only coexist but complement each other. The base-pair nucleotide sequence of DNA, the typical subject of study in classic genetics, cannot completely explain the observations that epigenetics seems to explain. These lessons are likely to lead us to reformulate our basic assumptions about the organization and role of the genome in phenotypic expression, evolution, and heredity[11].
The Origin of Disease
A number of studies have focused on genome- wide assays to indicate a possible cause and effect relationship with a disease and epigenetic changes. While we are tempted to conclude a cause and effect relationship, changes to epigenetic markers don’t necessarily cause disease, but may be a consequence of disease[12]. A study that is frequently used as an example of this conundrum involves people of high body mass index that had unusual epigenetic markers on a specific gene[13]. Initially, there was celebration among the researchers because they found a specific marker that could be manipulated and revolutionize the treatment to cure obesity. Their hopes were dashed when a subsequent study found that it was only after the people gained weight that the epigenetic marker appeared.[14]
Other studies that were more appropriately designed, showed a universal loss of the maternal allele in Wilms’ tumor. With loss of imprinting, an unidentified tumor suppressor gene appears to be expressed.[15] Loss of this specific allele has been found to be common in lung cancer, breast cancer, and ovarian cancer. The emergent challenge is to now find out how these changes in epigenetic markers might be harnessed to find a cure for cancer.
Effect on Behavior
Autism and bipolar disorder are two common complex traits that have defied gene identification, and both show surprisingly high frequencies of phenotypic discordance in identical twins, implying epigenetic changes to account for the differences.[16] Maternal behavior in rats can affect long-term behavior in their offspring and this correlates with changes in DNA methylation and histones attached to a glucocorticoid receptor gene promotor in the pup’s hippocampus. In the same study, the researchers found that the effects were not permanent; that in fact, giving the drug Reichstein A or the amino acid l-methionine to older pups could help reverse the behavioral effects of poor maternal care.[17] This is an example of what epigenetics may be able to offer for addressing the source of the problem.
Effect of Diet and Nutrition on Epigenetics
In general, mainstream medicine does not focus on nutrition but instead just advises patients to “eat a healthy diet”. Though animal evidence linking nutrition and DNA methylation is fairly extensive, comparable studies in humans are limited. In theory, anything that interferes with methylation is bound to have a profound effect on epigenetic expression. Methylation is dependent on an adequate supply of methyl donors, the major ones being S-adenosyl-methionine (SAM) and folate. SAM is not produced unless folic acid, Vitamin B12, and other nutritional co-factors are present such as methionine, betaine, and choline. A deficiency of any of these vitamins and cofactors can lead to hypomethylation.[18] The effects of a low folate diet can be reversed, but it can take months.
Peri-conception is a very vulnerable time for the developing fetus and deficiencies at this time can have effects on development and into adulthood. Flour is now fortified with folate to significantly reduce the incidence of neural tube defects[19]. In an animal study[20] using mature female sheep, restriction of folate, vitamin B-12, and methionine from the peri-conceptional diet-induced obesity in adult offspring as well as altered immune responses to an antigenic challenge. All of these changes were associated with a widespread decrease in methylation.
Environment and Stressors
Human behavior in adulthood is greatly influenced by various environmental stressors during childhood and adolescence.[21] To study this question, adolescent mice predisposed to neurological disorders were placed in solitary confinement from the age of 5 to 8 weeks and then returned to group housing after adolescence (from 8 to 20 weeks). At 8 and 20 weeks of age, they examined DNA methylation in the Th gene between the control and isolated group. The significantly high levels of methylation in the isolated animals were much greater when compared to the control group at 8 and 20 weeks. They concluded that isolation stress during adolescence can elicit molecular, neurochemical, and behavioral deficits when combined with an appropriate genetic risk.[22]
In another study[23], rats were stressed from gestational days 12 to 18 while others served as handled controls. Gestational stress in the mother disrupted post-partum maternal behavior and was accompanied by brain miRNA differences compared with control in both the mother and her offspring, and altered transcribed brain profiles in the offspring. Offspring gene expression changes included genes related to development, axonal guidance, and neuropathology. These findings indicate that prenatal stress modifies epigenetic signatures linked to disease during critical periods of fetal brain development. The authors concluded that these observations provide a new mechanistic association between environmental and genetic risk factors in psychiatric and neurological disease.
Environmental Toxins
A lot has been written about the influence of toxins on our health. And generally, we feel a sense of security when we know that the levels in our environment are in the “safe zone”. However, recent research demonstrates that endocrine disrupting chemicals (EDCs) at infinitesimally small amounts can wreak havoc on development and that these changes can be heritable for several generations. This means that the addition of any exogenous chemicals and/ or EDCs automatically exceed thresholds for affecting development. Rapidly growing evidence has linked environmental pollutants with epigenetic variations, including changes in DNA methylation, histone modifications, and microRNAs.[24]
A very elegant study was carried out by Diaz and Kessler[25] to demonstrate the effect of parental chemical exposure on the behavior of successive generations. Adult mice (F0) were subjected to odor fear conditioning with acetophenone, a chemical known to activate a specific odorant receptor in mice. The exposed mice were bred and the subsequent F1 and F2 generations showed an increased behavioral sensitivity to the F0-conditioned odor, but not to other odors. Even more importantly, genetic analysis reveals hypomethylation of the olfactory receptor gene that was stimulated in the F0 mice. Their findings provide a framework for addressing how the effects of environmental toxins may be inherited transgenerationally at behavioral, neuroanatomical and epigenetic levels.
Darwin versus Lamarck
Epigenetics represents a fatal flaw in Darwin’s theory of natural selection where DNA changes over time were relevant. Epigenetics has elements that hark back to the discredited theories of Jean-Baptiste Lamarck, who in the 1900’s proposed that organisms pass down acquired traits to future generations. That is where the similarities to epigenetics end. He also believed that giraffes have long necks because they had to reach the top of trees to get food and they passed this trait on to their young. At the turn of the 20th century, August Weismann debunked that theory by chopping off the tails of mice to prove that their pups could not inherit their tail-lessness.
Other Explanations
The mechanism of epigenetics has been attributed to methylation, histones, and RNA although there could be other explanations for the observations related to things outside our field of vision. These might include things such as prions,[26] viruses, or something as yet undiscovered. We are just now getting a hint at what epigenetics can do in the way of clinical applications while we sort through the evidence currently available. Clarifying the possibilities for clinical applications will keep scientists busy for quite some time.
Value of Epigenetics Now
Despite the fact that we are only touching the surface of epigenetics, sufficient information has been gathered and harnessed resulting in a number of clinically relevant applications. There is no field of medicine, biology, or sociology that is ignoring the opportunities made available by epigenetics and scientists are dreaming up novel ways to apply this new knowledge. As examples of what can be accomplished with our understanding of epigenetics, three areas will be explored: male infertility, stem cells, and cancer therapy.
Male Infertility
Spermatozoa have often been viewed simply as the motile means by which male DNA is delivered to the awaiting oocyte. In other words, sperm were thought to be passive. Current research takes issue with that assumption and as technology advances, it becomes clear that sperm are as involved in regulation of embryonic development as the egg is.[27] This introduction of new and contrasting data has moved the field to now ponder a new question for research; how does a terminally differentiated sperm prepare to make a totipotent embryo?
Understanding the details of the normal function of sperm has been like slowly peeling the layers of an onion. From genetic analysis to epigenetics to proteomics, new information has been gained as technology and knowledge appearing at an almost logarithmic rate has advanced. There are approximately 2700 genes involved in spermatogenesis.[28] As part of this process, spermatozoa undergo a complete remodeling and repackaging as they mature and prepare for fertilization. During the final stage of spermiogenesis, 90-95% of histones are stripped from the DNA and replaced with protamine.[29] The residual histones and mRNA appear to be necessary for post-fertilization paternal gene regulation of embryogenesis and maintenance of imprinted gene markers.[30]
DNA methylation is a potent inhibitor of gene expression. At the time of fertilization, most of the sperm DNA is de-methylated by an unknown process.[31] Between the removal of histones and de-methylation of DNA, most of the DNA is naked and thus available for transcription and embryogenesis. As development proceeds, DNA is again progressively methylated, as certain genes are no longer required.[32]
Having some clues as to how normal sperm function opens windows into understanding the mechanisms of male infertility. Issues such as aging of the prospective father (discussed above), using abnormal sperm to fertilize an egg using ICSI, sperm preservation, and sperm donation all have in common the risk of paternal inheritance of abnormal genetic or epigenetic information.[33] Some of the issues already identified are a small increased risk of imprinting disorders in offspring of ICSI[34], abnormal methylation patterns in male infertility[35], abnormal histone modifications in infertility[36], and altered amounts of protamine in sperm associated with infertility and abnormal embryo development.[37]
Based on the methylation and histone modifications that have been discovered in sperm, it is now possible to predict male fertility. Episona is a molecular information company that develops personalized diagnostics, based on the science of epigenetics. Our first product, Seed, is the first diagnostic test to predict male infertility by analyzing epigenetic modifications at the level of the DNA, making it easier to guide treatment decisions. This association between epigenetic profile and infertility exists even when the semen parameters (i.e., count, morphology, and motility) are normal.
Cancer Research
Male fertility is just one of the spin-offs from epigenetic technology. The DNA-methylation and histone-modification patterns associated with the development and progression of cancer have potential clinical use. DNA hypermethylation markers are under study as diagnostic tools, prognostic factors, and predictors of responses to treatment. Unlike mutations, DNA methylation and histone modifications are reversible. It is possible to re-express DNA-methylated genes in cancer cell lines by using demethylating drugs, two of which appear to have some effect at very low doses. Drugs that deactivate the changes to histones are also being tried in vitro[38]. One concern about using these drugs is that they are nonspecific and could have unintended consequences in normal tissues. In addition to anti-cancer drugs, other chemicals are being assessed for their ability to selectively alter epigenetics. Other markers have added to our knowledge of the cells of origin of cancer[39] which will aid in personalized treatment.
Cell Reprogramming
Stem cells have excited scientists and physicians because of the great potential for gene transfer, restoring damaged tissue, in vitro drug testing, modeling diseases, and reversing the ravages of old age. At this time there are 2 sources of stem cells: embryonic stem cells (ES) and human somatic cells. The former is difficult to acquire and the latter has a low yield from reprogramming efforts. Induced pluripotent cells (iPS) are a third option, slowing moving from animal models to human application.
The significance of iPS cannot be overstated in terms of research and application in this area; it appears to be the best alternative because the needed stem cells are derived from the patient’s own somatic cells that are cultured in a media containing 3 or 4 transcription factors. In this technique in mice, it produces a high yield of stem cells that appear to be equivalent to ES in gene expression and embryo development. In humans, the gene expression is similar, but there are some differences in iPS and ES clones and cell lines. Studies are underway to find out just what the differences are and whether they are relevant to clinical trials with iPS.[40]
The ability to reprogram cells has resulted in a race to further define what can be done with reprogramming ability and potential clinical applications. The possibilities are limitless and the future of reprogramming cells is bright, specifically due to the scientific exploration now occurring in this fascinating emergent field.
[1] Waddington, C.H. The epigenotype. Endeavour, 1942; 1, 18–20.
[2] https://scienceblogs.com/pharyngula/2008/07/22/epigenetics/
[3]Guerrero-Bosagna. High Type II Error and Interpretation Inconsistencies When Attempting to Refute Transgenerational Epigenetic Inheritance. Genome Biology, 2016; 17:153.
[4] https://www.usc.edu/hsc/info/pr/ccr/07winter/epigenetics.html
[5] Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;287-301.
[6] Pfeiffer K. Mechanisms of genetic imprinting. American Journal of Human Genetics. 2000 Oct; 67(4): 777–787.
[7] Smith ZD, et al. DNA Methylation dynamics of the human preimplantation embryo. 2014 July; 511: 611–615.
[8] Crick FHC. Central Dogma of Molecular Biology, Nature, 1970;227:561-563.
[9] Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reproductive Toxicology 2005; 20:345–352.
[10] Park JH, Withholder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Natural Genetics 2010; 42:570–5.
[11] Shapiro JA. Revisiting the central dogma in the 21st century. Natural Genetic Engineering and Natural Genome Editing: Annals of the New York Academy of. Science. 2009;1178: 6–28.
[12] Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004; 429:457-63.
[13] Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014; 383:1990–1998.
[14] Richmond RC, Sharp GC, Ward ME, et al. DNA methylation and body mass index: investigating identified methylation sites at HIF3A in a causal framework. Diabetes. 2016; db150996.
[15] Steenman MJ, Rainier S, Dobry CJ, et al. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumor. Nature genetics. 1994; 7:433-9.
[16] Crews, DC, Gore AC. Transgenerational epigenetics: current controversies and debates. Transgenerational Epigenetics: Evidence and Debate. Amsterdam: Elsevier (2014): 371-91.
[17] Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nature neuroscience. 2004; 7:847-54.
[18] Jacob RA, Gretz DM, Taylor PC, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. Journal of Nutrition, 1998;128:1204–12.
[19] Honein MA, Paulozzi LJ, Mathews TJ, et al. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. Journal of the American Medical Association. 2001;20;285:2981-6.
[20] Sinclair KD, Allegrucci C, Singh R, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal peri-conceptual B vitamin and methionine status. Proceeding of the National Academy of Science USA. 2007; 104:19351–6.
[21] Blakemore SJ. The social brain in adolescence. Nature Reviews Neuroscience. 2008; 9:267-77.
[22] Niwa M, Jaaro-Peled H, Tankou S, et al. Adolescent stress–induced epigenetic control of dopaminergic neurons via glucocorticoids. Science. 2013; 339:335-9.
[23] Zucchi FC, Yao Y, Ward ID, et al. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PloS one. 2013 Feb 22;8(2):e56967.
[24]Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature neuroscience. 2014; 17:89-96.
[25] Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007 Feb 23;128(4):655-68.
[26] Trivedi BP. Outside the fold. Nature 2012; 482:294-296.
[27]Carrell DT, Aston1 KI, Oliva R, et al. The “omics” of human male infertility: integrating big data in a systems biology approach. Cell Tissue Res (2016) 363:295–312. DOI et al 10.1007/s00441-015-2320-7
[28] de Mateo S, Martinez-Heredia J, Estanyol JM, et al. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 2007;7:4264–4277.
[29] Aoki VW, Carrell DT. Human protamines and the developing spermatid: their structure, function, expression and relationship with male infertility. Asian J Androl 2003;5:315–324.
[30] Dunn GA, Morgan CP, Bale TL. Sex-specificity in transgenerational epigenetic programming. Horm Behav 2013;59:290–295.
[31] Erkek S, Hisano M, Liang CY, et al. Molecular determinants of nucleosome retention at CpG-rich sequences inmouse spermatozoa. Nat Struct Mol Biol 2013;20:868–875.
[32] Borgel J, Guibert S, Li Y, et al. (2010) Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 2010;42:1093–1100.
[33] Morris ID, Ilott S, Dixon L, Brison DR. The spectrum of DNA damage in human sperm assessed by single cell gel electrophoresis (Comet assay) and its relationship to fertilization and embryo development. Human Reproduction. 2002;17(4):990-8.
[34] Lazaraviciute G, Kauser M, Bhattacharya S, Haggarty P, Bhattacharya S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Human reproduction update. 2014;Jun 24:dmu 033.
[35] Aston KI, Punj V, Liu L, Carrell DT. Genome-wide sperm deoxyribonucleic acid methylation is altered in some men with abnormal chromatin packaging or poor in vitro fertilization embryogenesis. Fertil Steril 2012;97:285–292.
[36] Hammoud SS, Nix DA, Hammoud AO, et al. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod 2011;26: 2558–2569.
[37] Aoki VW, Emery BR, Liu L, Carrell DT. Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl 2006;27:890–898.
[38] Esteller M. Epigenetics in cancer. New England Journal of Medicine. 2008; 58:1148-59.
[39] Visvader JE. Cells of origin in cancer. Nature. 2011;469:314-22.
[40] Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295-305.